Don’t overinterpret STRUCTURE plots
Several weeks ago1 Daniel Falush (@DanielFalush) posted a preprint on bioRxiv, “A tutorial on how (not) to over-interpret STRUCTURE/ADMIXTURE bar plots”. I finally had a chance to read it this weekend. Here’s the abstract:
Genetic clustering algorithms, implemented in popular programs such as STRUCTURE and ADMIXTURE, have been used extensively in the characterisation of individuals and populations based on genetic data. A successful example is reconstruction of the genetic history of African Americans who are a product of recent admixture between highly differentiated populations. Histories can also be reconstructed using the same procedure for groups which do not have admixture in their recent history, where recent genetic drift is strong or that deviate in other ways from the underlying inference model. Unfortunately, such histories can be misleading. We have implemented an approach (available at www.paintmychromsomes.com) to assessing the goodness of fit of the model using the ancestry ‘palettes’ estimated by CHROMOPAINTER and apply it to both simulated and real examples. Combining these complementary analyses with additional methods that are designed to test specific hypothesis allows a richer and more robust analysis of recent demographic history based on genetic data.
A key observation Falush and his co-authors make is that different demographic scenarios can lead to the same STRUCTURE diagram. They illustrate three different scenarios. In all of them, they simulate data from 12 populations but sample from only four of them. In all of the scenarios, population P4 has been isolated from the other three populations in the sample for a long time. It’s the relationship between P1, P2, and P3 that differs among the scenarios.
- Recent admixture: P1 and P3 have also been distinct for some time, and P2 is a recent admixture of P1, P3, and P4.
- Ghost admixture: P1 and P3 diverged some time ago, and P2 is a recent admixture of P1 and a “ghost” population more closely related to P3 than to P1.
- Recent bottleneck: P1 is sister to P2 but underwent a strong recent bottleneck.
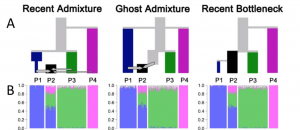
As you can see, the STRUCTURE diagrams estimated from data simulated in each scenario are indistinguishable. They also show that if you have additional data available, specifically if you are lucky enough to be working in an organism with a lot of SNPs that are mapped, then you can combine estimates from CHROMOPAINTER with those from STRUCTURE to distinguish the recent admixture scenario from the other two – assuming that you’ve picked a reasonable number for K, the number of subpopulations.2
The authors also refer to Puechmaille’s recent work demonstrating that estimates of genetic structure are greatly affected by sample size. Bottom line: Read both this paper and Puechmaille’s if you use STRUCTURE, tread cautiously when interpreting results, and don’t expend too much effort trying to estimate the “right” K.
1OK, as you can see from the tweet, it was almost a month ago.
2The paper contains a brief remark about how hard it is to estimate K: “Unless the demographic history of the sample is particularly simple, the value of K inferred according to any statistically sensible criterion is likely to be smaller than the number of distinct drift events that have significantly impacted the sample. What the algorithm often does is in practice use variation in admixture proportions between individuals to approximately mimic the effect of more than K distinct drift events without estimating ancestral populations corresponding to each one.”
Falush, D., L. van Dorp, D. Lawson. 2016. A tutorial on how (not) to over-interpret STRUCTURE/ADMIXTURE bar plots. bioRxiv doi: 10.1101/066431
Lawson, D.J., G. Hellenthal, S. Myers, and D. Falush. 2012. Inference of population structure using dense haplotype data. PLoS Genetics 8:e1002453. doi: 10.1371/journal.pgen.1002453
Puechmaille, S.J. 2016. The program structure does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Molecular Ecology Resources 16:608-627. doi: 10.1111/1755-0998.12512


 National Geographic has made available nearly all of the topographic quadrangle maps from the US Geological Survey as PDF download.
National Geographic has made available nearly all of the topographic quadrangle maps from the US Geological Survey as PDF download. In early June, Michael Gove was Secretary of State for Justice and Lord Chancellor of the United Kingdom. (Image at left By Policy Exchange [CC BY 2.0 (
In early June, Michael Gove was Secretary of State for Justice and Lord Chancellor of the United Kingdom. (Image at left By Policy Exchange [CC BY 2.0 (